Source Genomics can offer analysis of different RNA Species (coding, non-coding and small transcripts) from a lengthy range of starting material using long and short read sequencing. We provide solutions for a wide variety of RNA Sequencing techniques. Offering excellent quality services with a fast turnaround time.
Total RNA sequencing
Total RNA sequencing involves the sequencing all the RNA within a sample. Typically, ribosomal RNA is removed from a sample, as this RNA is highly abundant and is non-coding. By sequencing the total RNA, we can look not only at the protein coding messenger RNA, but also the non-coding RNA transcripts that may be involved in interference and regulation of different genes.
Transcriptome sequencing
Transcriptome sequencing involves sequencing the mRNA, the RNA which directly codes for proteins. By sequencing the mRNA, it is possible to identify which genes are over-and-under expressed, as well as identifying novel transcripts within the transcriptome. We also provide De novo transcriptome assembly for organisms previously not sequenced.
Small RNA sequencing
Small RNA sequencing utilises a preparation which selects for small RNAs within a sample. By sequencing small RNAs, and analysing which are expressed, the presence of interfering molecules can be identified, which are an important for post-transcriptional gene silencing and regulation.
Sc (single cell) RNA Sequencing
Single cell sequencing provides a higher resolution of cellular differences and a better understanding of the transcriptome of an individual cell in the context of its micro-environment.
We provide run only options leaving you the flexibility to choose the library preparation supplier of your choice, to best suit your needs. Additionally, we can support library preparation from 10x Genomics experiments from cDNA generation.
A free consultation is provided at the initiation stage of the project and an a dedicated Account Manager will be assigned to provide end-to-end support throughout the project and post data delivery.
Overview
Sequencing package details
At Source Genomics we realise that the standard package does not always fit the customer needs – we offer the flexibility to increase/decrease both the number of reads and read length required. Our standard package includes:
• Initial QC assessment of RNA
• Library preparation from as little as 2pg total RNA
• Sequencing to a depth of 30 million clusters with PE150bp reads
• Demultiplex and return of data
RNA extraction from various sample sources as well as advanced Bio-informatic analysis is also available to support your entire project needs.
The process
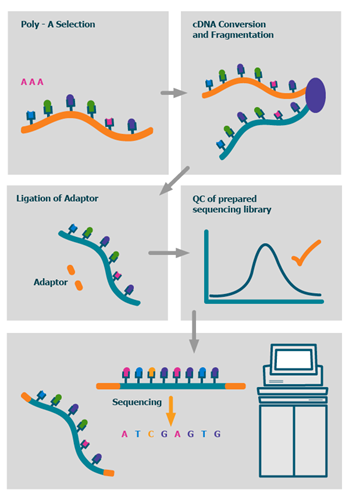
We also offer advanced BioIT variant analysis packages as well as fully bespoke BioIT solutions.
BioInformatics
Digital gene expression
Quantification of transcripts in a sample using RNA-Seq data
This analysis uses RNA-Seq data to quantify expression levels by comparison to a reference genome, or de novo transcriptome assembly if a reference genome is not available. Raw reads are trimmed, aligned and quantified by gene-wise read counting using the featureCounts model. You will receive raw (fastq) and aligned (bam) files for each sample as well as text files detailing transcripts per million (TPM) or fragments per kb per million mapped reads (FPKM). A bioinformatics report will provide further detail about the tools used, alignment metrics and a summary of results.
Differential gene expression
Pairwise comparison of gene expression between sample groups
This analysis is similar to digital gene expression, but includes a comparative analysis between groups of samples. You will receive raw (fastq), aligned (bam) files and text files of gene counts as well as a PCA plot of all samples, a sample-to-sample distance heatmap and MA plots and differential expression heatmaps for each comparison. A bioinformatics report will provide further detail about the tools used, alignment metrics and a summary of results.
Other gene expression methods
Bespoke analyses of other RNA-Seq data, such as (differential) miRNA or methylation quantification; identification of splice variants; detection and quantification of fusion genes.
Transcriptome assembly
De novo or reference-guided assembly of RNAseq data into one transcriptome
Depending on the availability of a reference sequence, reads are de novo assembled, or against a reference genome (or a combination of the two) to create sequence scaffolds.
The analysis includes quality trimming of raw reads, assembly into contigs – mapping against a reference, if available – filtering and creating sequence scaffolds. You will receive raw (fastq) files, the assembled sequence file (fasta) and a bioinformatics report detailing the tools used and contig size statistics.
How to order
Contact us today and one of our skilled account managers will be in touch with a free consultation including further information and pricing details.
Payment options:
Payment can be made by credit card or purchase order number.
FAQ's:
View our frequently asked questions for more information.
Sample requirements:
We require 1-2 ug of gDNA (0D260/280 ratio ranging from 1.8-2) at a minimum concentration of 50 ng/μl and in a minimum volume of 20 μl. Higher concentrations can be submitted and will be diluted accordingly. For full details click here.
How to order
Contact us today and one of our skilled account managers will be in touch with a free consultation including further information and pricing details.
Next Generation Sequencing
Source BioScience is one of Europe’s leading providers of commercial sequencing, offering Next Generation Sequencing services from our ISO accredited laboratories. We offer NGS services on the most prominent platforms including Illumina’s NovaSeq, NextSeq and MiSeq.